Handheld and fully integrated 1-channel current amplifier for ultra-low-noise measurements. Suitable for lipid bilayer membrane studies.
Nanopore Reader 100 KHz
BLM Chips for protein reconstitution
BLM Chip MM
A microfluidic system serving as the interface between the bilayer and the current reader. Designed for Montal-Müller technique.
Lipid Bilayer Formation: Vertical
Chambers Volume: 80 µl
Aperture Sizes: 50 ; 75 ; 100 ; 140 µm
Typical Membrane Capacitance (DPhPC-made lipid membranes; 25 µm hole thickness): 20-40 pF in 50 µm, 40-60 pF in 75 µm, and 60-90 pF in 100 µm, 90-140 pF in 140 µm
Replaceable External Silver Wires
Reusable Chip (up to 30 times)

BLM Chip Standard
A microfluidic system serving as the interface between the bilayer and the current reader. Designed for pseudo-bubble method and paint brush painting.
Lipid Bilayer Formation: Horizontal
Chambers Volume: 60 µl
Aperture Sizes: 100 ; 150 µm
Typical Membrane Capacitance (DPhPC-made lipid membranes): 20-50 pF in 100 µm, and 50-110 pF in 150 µm.
Cis and trans compartments accessible from top side
Non Replaceable Printed Silver Electrodes
Reusable Chip
Use Cases
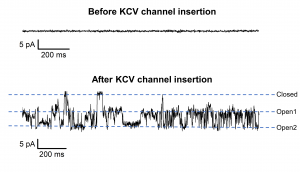
Exemplary current trace of KCVnts channel recorded at -60mV in symmetrical 1M KCl, PH 7 (with 1,25 KHz sampling rate and 625 Hz bandwidth). The channel protein was translated in vitro into NDs (nanodiscs) with DMPC membranes. The purified NDs in dilution with imidazole were directly administered to the bilayer formed by painting DPhPc over our 150 µm diameter BLMchip designed for biological pores. The open and closed level of the channel are indicated. The top panel shows the signal of the membrane at -60mV before the insertion of the channel protein. – Prof. G. Thiel (TU-Darmstadt University).
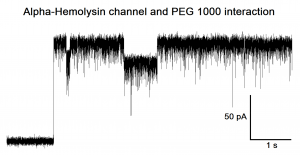 Exemplary current trace of a single α-Hemolysin channel insertion recorded at 5 KHz sampling rate (and 2,5 KHz bandwidth), applying a membrane potential of 100mV, in symmetrical 1M KCl, PH 7. PEG1000 molecules (already present in solution) translocate through the α-Hemolysin channel giving rise to negative individual blockades. The channel protein was directly added in solution and auto-assembled into the bilayers made of DPhPc.
Exemplary current trace of a single α-Hemolysin channel insertion recorded at 5 KHz sampling rate (and 2,5 KHz bandwidth), applying a membrane potential of 100mV, in symmetrical 1M KCl, PH 7. PEG1000 molecules (already present in solution) translocate through the α-Hemolysin channel giving rise to negative individual blockades. The channel protein was directly added in solution and auto-assembled into the bilayers made of DPhPc.
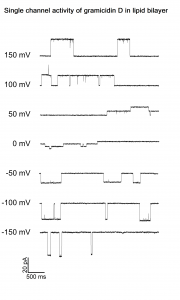 Exemplary current trace of gramicidin D single channel. A bilayer membrane was formed by painting DPHPC lipids (10mg/ml in n-octane) over the 100uM hole of the BLM-chip filled with asymmetrical HCl solutions. When a stable membrane was obtained, a small amount of gramicidin was added to the recording solutions. After some minutes, the formation of gramicidin dimers allowed the flow of H+ ions across the membrane.
Exemplary current trace of gramicidin D single channel. A bilayer membrane was formed by painting DPHPC lipids (10mg/ml in n-octane) over the 100uM hole of the BLM-chip filled with asymmetrical HCl solutions. When a stable membrane was obtained, a small amount of gramicidin was added to the recording solutions. After some minutes, the formation of gramicidin dimers allowed the flow of H+ ions across the membrane.
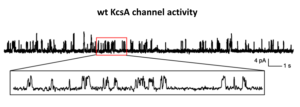 Representative single channel currents of wt KcsA reconstituted in liposomes and incorporated in POPE:POPG lipid bilayers (w:w, 3:1).
Representative single channel currents of wt KcsA reconstituted in liposomes and incorporated in POPE:POPG lipid bilayers (w:w, 3:1).
– Data courtesy of Prof. M. F. Tsai (University of Colorado School of Medicine, USA).
 Representative current trace showing single channel activity of chloride intracellular channel 1 (CLIC1) reconstituted in DPhPC lipid bilayer membranes painted over a 150 µm hole.
Representative current trace showing single channel activity of chloride intracellular channel 1 (CLIC1) reconstituted in DPhPC lipid bilayer membranes painted over a 150 µm hole.
– Data courtesy of Prof. M. Mazzanti (University of Milan, Italy).
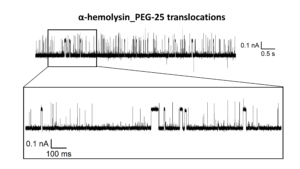 PEG-25 (10 µM) molecules translocation through a single α-Hemolysin nanopore recorded at -100 mV, 20kHz SR (10 kHz BW). The protein was inserted into a DPhPC-made lipid bilayer membrane painted into our BLMchip embedding a 150 µm sized hole. The recording solution contained 3M KCl, 20mM TRIS, PH 8.
PEG-25 (10 µM) molecules translocation through a single α-Hemolysin nanopore recorded at -100 mV, 20kHz SR (10 kHz BW). The protein was inserted into a DPhPC-made lipid bilayer membrane painted into our BLMchip embedding a 150 µm sized hole. The recording solution contained 3M KCl, 20mM TRIS, PH 8.
Your Lipid Bilayer Experiment using the BLM Chip
Nanopore Reader 100 kHz Noise Levels
- Open input (RMS) noise (Voltage range ±700mV) : 0.06 pA rms @ 625Hz; 0.3 pA rms @ 10 kHz; 2.4 pA rms @ 100 kHz
- Open input (RMS) noise (Voltage range ±2000mV) : 0.08 pA rms @ 1kHz; 0.42 pA rms @ 10 kHz; 3.7 pA rms @ 100 kHz
- Current ranges: ±200pA (Gain 2.25GΩ), ±2nA (Gain 225MΩ), ±20nA (Gain 22.5MΩ), ±200nA (Gain 2.25MΩ)
- Voltage hold ranges: ±700mV (ultra low noise); ±2000mV (low noise)
- Parametric voltage protocols
- Max sampling rate: 200 ksps
- Selectable x4 oversampling (max final sampling rate 800 ksps)
- Available bandwidth between 625 Hz to 100 kHz
- Auto electrodes voltage offset fine compensation
- Continuous Capacitance and Resistance estimation
- USB powered
- Size & Weight: 101 x 44 x 18 mm, 140 g
User Guides
- Connection diagram
- Get started with the model cell
- Ultra low noise modality
- Nanopore Reader Voltage protocols
- How to use the Nanopore Reader adaptor to connect external recording chamber
- How to prevent damage to Nanopore Reader Amplifiers – Best Practice for flow cell handling
- ESD Handling Guidelines for Elements Amplifiers
Technical Guides
Biological Nanopores
Solid State Nanopores
- Multimodal nanoparticle analysis enabled by a polymer electrolyte nanopore combined with nanoimpact electrochemistry, Gyasi Agyemang et al., Faraday Discussions, 2024
- High Accuracy Protein Identification: Fusion of Solid-State Nanopore Sensing and Machine Learning, Dutt, S., Shao, H., Karawdeniya, B., Bandara, Y. M. N. D. Y., Daskalaki, E., Suominen, H., Kluth, P., Small Methods 2023, 2300676
- Ultrathin, High-Lifetime Silicon Nitride Membranes for Nanopore Sensing, Dutt S et al., Anal. Chem., 2023
- Large-scale production of polyimide micropore-based flow cells for detecting nano-sized particles in fluids, Salehirozveh et al., RSC Adv., 2023, 13, 873-880
- Nanopores: a versatile tool to study protein dynamics, Schmid S, Dekker C., Essays Biochem., 2021
- Detection of single analyte and environmental samples with silicon nitride nanopores: Antarctic dirt particulates and DNA in artificial seawater., Niedzwiecki DJ et al., Rev Sci Instrum., 2020 Mar 1;91(3):031301.
- DNA fragment translocation in artificial sea water through nanopores using a portable mini reader and flow-cell., Niedzwiecki DJ et al., Poster presented at BPS meeting.

Custom device development
With our custom ASIC design technology we can help you configure specific tools and solutions for your applications.
Let us know how we can help you design the tools and software you need.




